Research
Enabling tools for proteomics
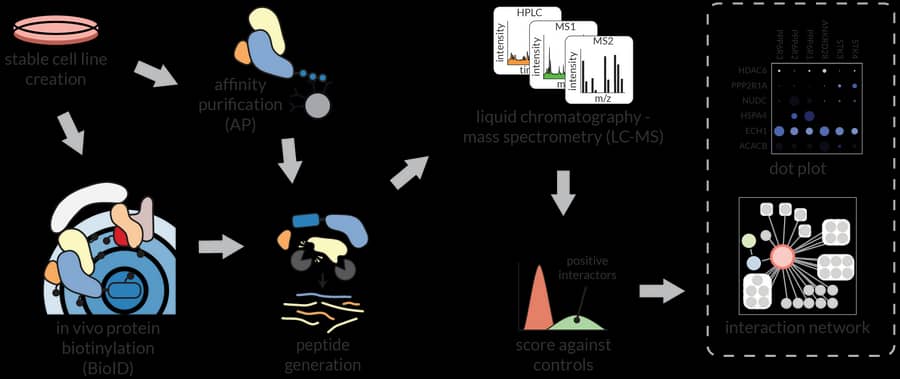
We are developing experimental and computational pipelines for proteomics, with a focus on interaction and affinity workflows. Experimentally, we have developed new sets of lentiviral vectors to express BioID fusions across multiple cell types (Samavarchi-Tehrani, MCP 2018), and protocols to perform proximity-dependent biotinylation in zebrafish embryos (Rosenthal, MCP 2021; with Ian Scott). Our current projects explore combining proximity-dependent biotinylation with in vivo crosslinking, looking at context-dependency with split enzymes, and developing approaches to perform proximity-dependent biotinylation extracellularly. Our computational tools range from a LIMS for proteomics (ProHits: Liu, Nature Biotechnology 2010) to tools for scoring protein-protein interactions (SAINT: Choi, Nature Methods 2011; CRAPome: Mellacheruvu, Nature Methods 2013), co-developed with Alexey Nesvizhskii, Hyungwon Choi and Mike Tyers. We also developed tools to visualize and further analyze interactions, most notably ProHits-viz (Knight, Nature Methods 2017), and introduced GIX (Knight, Nature Methods 2019), a browser extension for rapid annotation of gene products. With Alexey Nesvizhskii, Nuno Bandeira and Hannes Röst, we have contributed to the development of tools for the interpretation of Data Independent Acquisition results. Current efforts aim to improve the connectivity between all these computational tools.
Systems biology
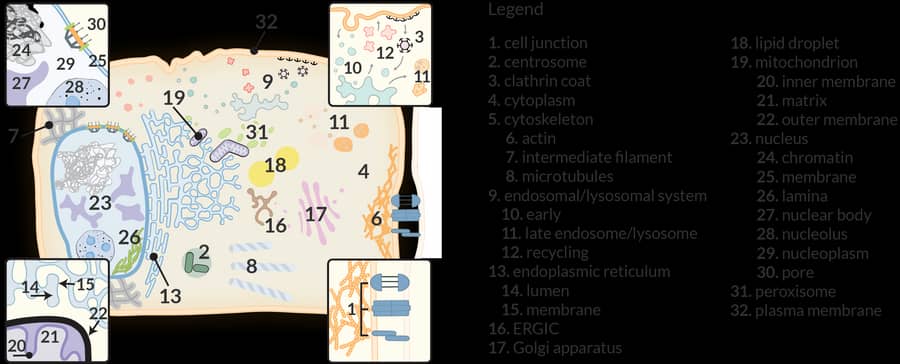
We have long taken a protein family-centric approach to explore specificity in the interactions established by kinases (Breitkreutz, Science 2010; with Mike Tyers and Alexey Nesvizhskii), phosphatases (St-Denis, Cell Reports 2016), bromodomain-containing proteins (Lambert, Molecular Cell 2018), and chaperones (Taipale, Cell 2014; Piette, Molecular Cell 2021). The introduction of proximity-dependent biotinylation in 2012 has now enabled us to provide an organelle-specific view of the proteome. We have notably described the proteome of cytosolic RNA-containing granules and bodies using BioID (Youn, Molecular Cell 2018; Youn, Molecular Cell 2019), and more recently, in collaboration with Eric Shoubridge, the organization of the human mitochondria (Antonicka, Cell Metabolism 2020). Lessons learned from these organelle-specific studies have enabled the establishment of a comprehensive proximity map of a human cell (Go, Knight, Nature 2021) and a dedicated website that enables user to navigate our results and analyze their own BioID experiments (humancellmap.org). We have also repurposed the humancellmap.org to help provide context to the proximity interactomes of SARS-CoV-2 proteins (Samavarchi-Tehrani, bioRχiv 2021; covid19interactome.org). Some of the current projects in the lab involve high-resolution maps of nuclear bodies, exploration of dynamic changes in the proximal proteomes following induction of somatic cell reprogramming, and studying the consequences of alternative splicing on interactomes.
Signalling
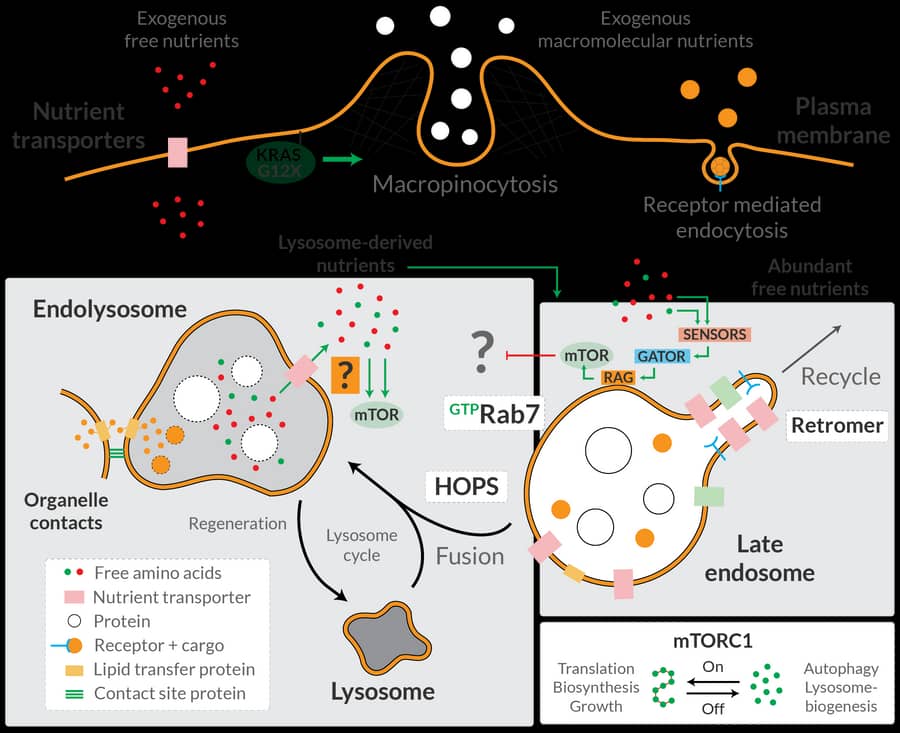
Some of our core research interests have been the study of signaling pathways that are deregulated in cancers and rare diseases. In 2009, we identified the STRIPAK complex (STRiatin-Interacting Phosphatase And Kinase; Goudreault, MCP 2009) that plays important roles in the Hippo signaling pathway (Couzens, Science Signaling 2013) and in Cerebral Cavernous Malformations, a vascular disease. In collaboration with the Sicheri lab, we have defined the phospho-dependent recruitment of proteins in the core kinase cascade of the Hippo pathway (Xiong, MCP 2017; Couzens, MCP 2017), and have helped our Terry Fox Program co-investigators to identify additional components in this pathway. Our efforts on the context-dependent aspects of Cerebral Cavernous Malformation signaling will make use of our newly developed in vivo zebrafish biotinylation system. Through the use of proximity-dependent biotinylation sensors, we have uncovered that the free-amino acid sensing pathway for mTORC1 activation (the GATOR-Rag GTPase pathway) actively suppresses an alternative pathway that activates mTORC1 through amino acids produced from lysosomal degradation of proteins (Hesketh, Science 2020). Future work in this area includes a more comprehensive mapping of the mTORC1 network, as well as continued efforts, some with the labs of Jean-François Côté and Matthew Smith, in characterizing proximity networks for small GTPases. Additionally, we are also exploring receptor kinase signaling using proximity-dependent biotinylation, looking at both intracellular and extracellular interactions.
COVID-19 serology
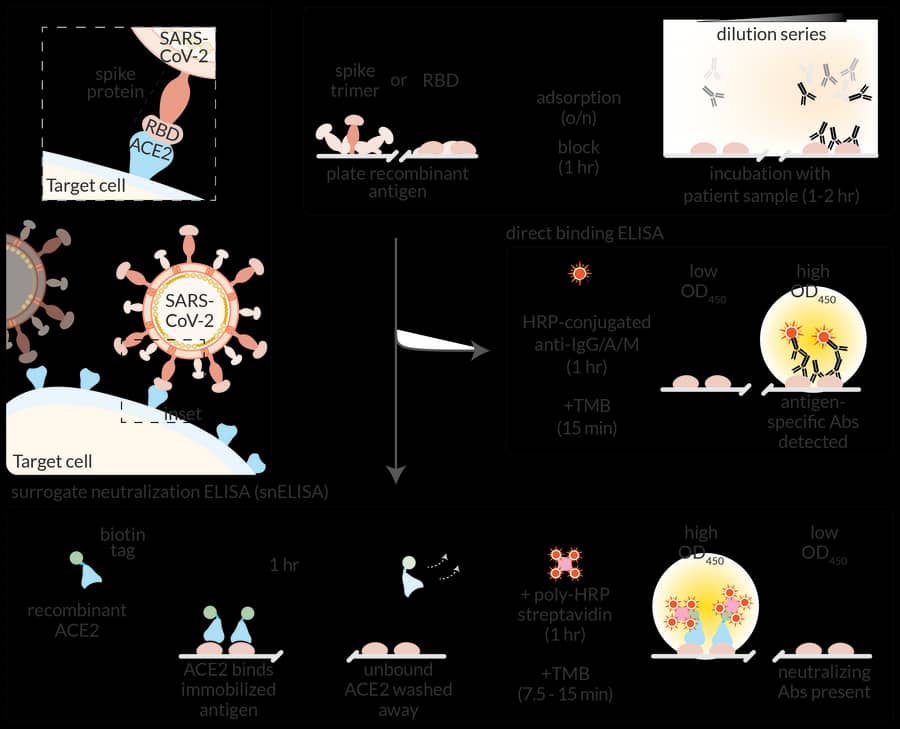
While the focus of the lab remains functional proteomics, systems biology and signal transduction, our expertise in protein-protein interactions and our affiliation with the Network Biology Collaborative Centre (NBCC) placed us in an excellent position to develop antibody-based assays for COVID-19 in March 2020. We received early funding through the Sinai Foundation, from the Krembil Foundation and the RBC Foundation that enabled us to build assays. With Jim Rini, we developed a protein-based neutralization surrogate for SARS-CoV-2 (Abe, JCI Insight 2020) and re-implemented a lentiviral neutralization surrogate assay introduced by Jesse Bloom. We partnered with colleagues at the National Research Council of Canada (NRC), the National Microbiology Laboratory (NML), colleagues in the Toronto Invasive Bacterial Diseases Network (TIBDN) and the Canadian Blood Services to develop high-throughput serology assays at the NBCC. These assays have been developed by a dedicated team, co-led by Anne-Claude and NBCC manager Karen Colwill. The assays have been used notably to demonstrate, in a collaboration with Jen Gommerman and Allison McGeer, that anti-spike antibodies in the blood and saliva of infected individuals lasted for at least 3-4 months (the latest time point collected in that particular study; Isho, Science Immunology 2020). The assays are now used for >40 different projects across the country that monitor seroprevalence or antibody levels following infection or vaccination, many funded through the Canadian Institutes of Health Research (CIHR) and the COVID-19 Immunity Task Force (CITF). Several of the studies in vaccinated populations focus on individuals with compromised immune systems, including residents of long-term care homes (with Sharon Straus and Allison McGeer), patients receiving dialysis (with Michelle Hladunewich), older individuals (with Sharon Walmsley), and patients with autoimmune disease (with Vinod Chandran, Tania Watts, Mark Silverberg, Sasha Bernatsky and Dawn Bowdish). We also contribute to development of new therapeutics and vaccines (see, e.g. Liu, Science Advances 2021). Lastly, Anne-Claude leads the Pillar 3: Functional Genomics and Structure/Function group for the newly funded CIHR network for Variants of Concern (CoVaRR-Net), led by Marc-André Langlois at the University of Ottawa.
Funding
We are grateful to the following agencies for supporting the projects in the Gingras lab and the LTRI proteomics group:
- Canadian Institutes of Health Research
- Natural Sciences and Engineering Research Council
- Canadian Cancer Society Research Institute
- Cancer Research Society
- Genome Canada
- Ontario Genomics Institute
- Canada Foundation for Innovation
- National Institutes of Health
- CIHR Institute of Genetics
- Compute Canada
- Canada Research Chair